A simple Soil - Target model Fixed individuals
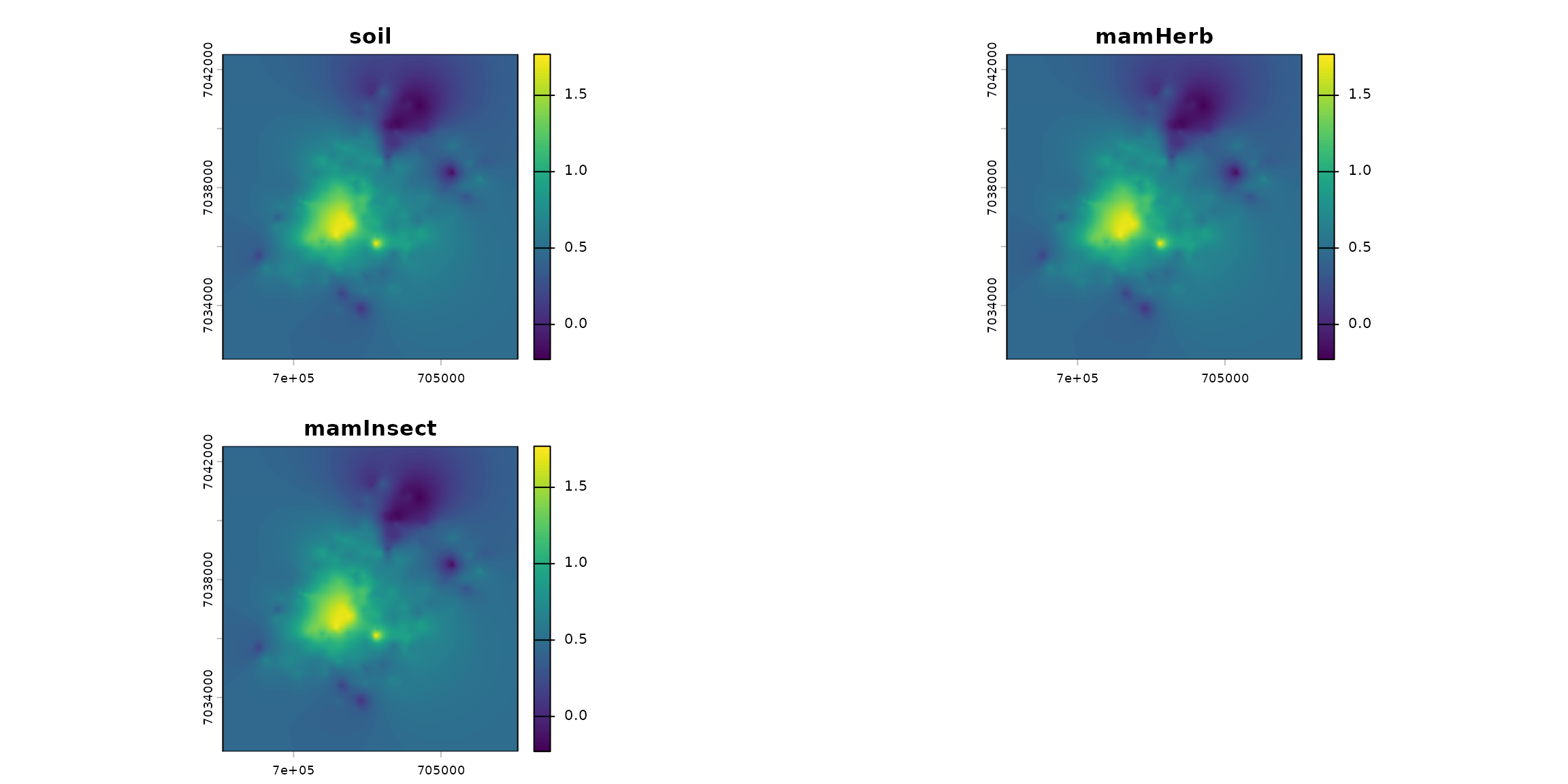
ground_cd <- load_raster_extdata("ground_concentration_cd_compressed.tif")
# names_hab = c("soil", "plant", "invert", "mamHerb", "mamInsect", "birdInsect")
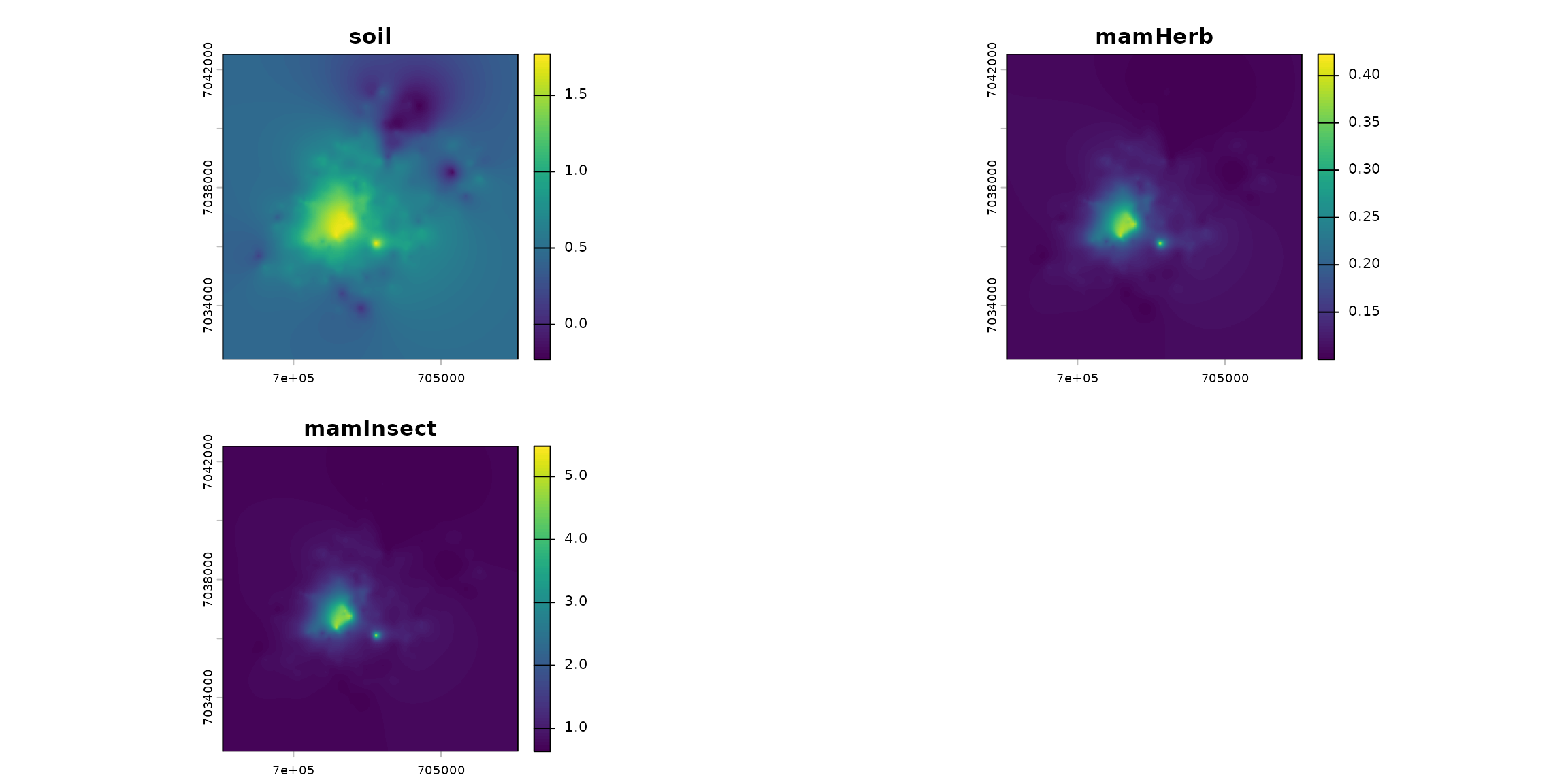
names_hab = c("soil", "mamHerb", "mamInsect")
list_habitat <- lapply(names_hab, function(i) ground_cd)
stack_habitat <- raster_stack(list_habitat, names_hab)
terra::plot(stack_habitat)
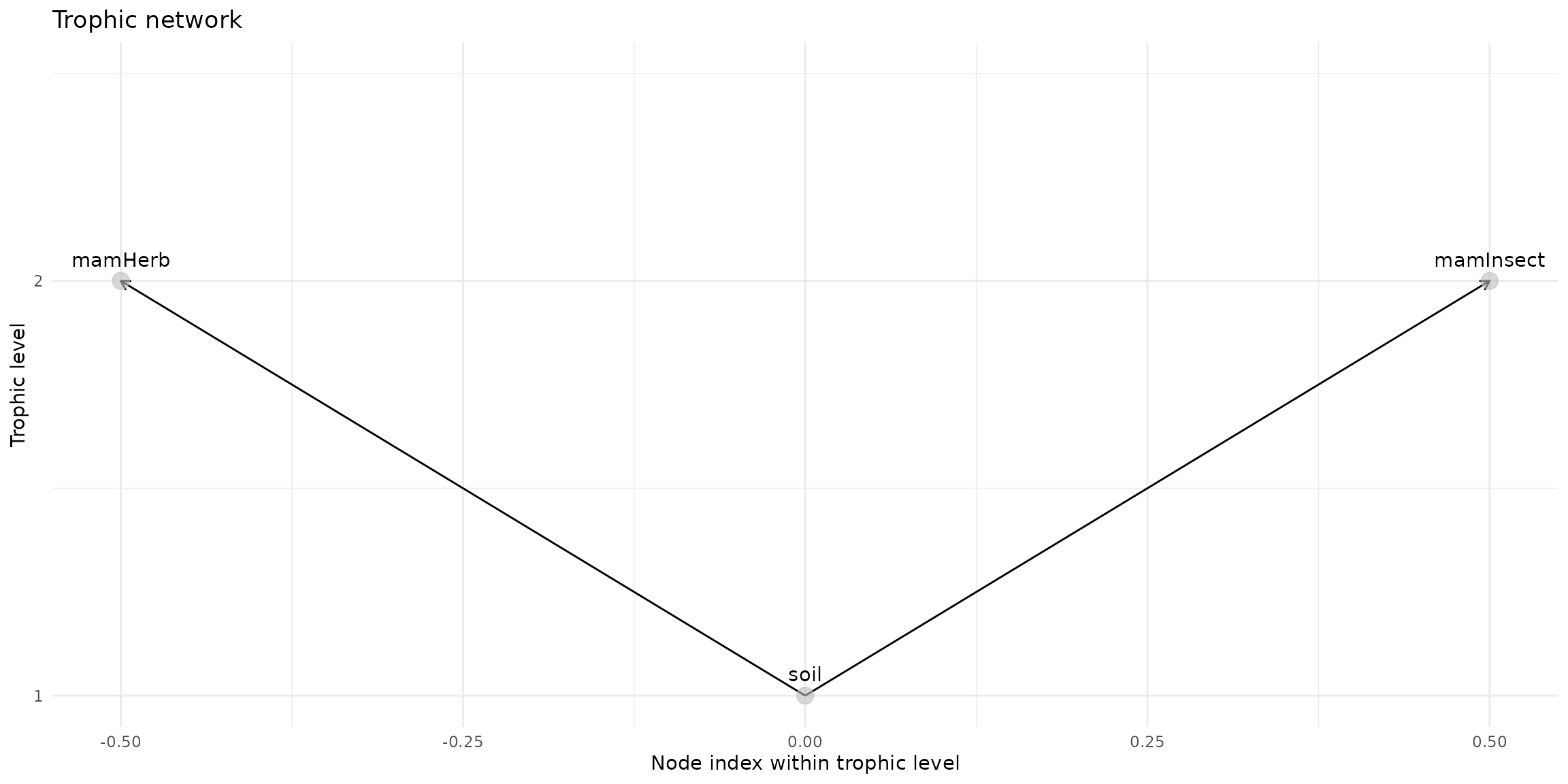
trophic_df <- trophic() |>
#add_link("soil", "plant") |>
#add_link("soil", "invert") |>
add_link("soil", "mamHerb") |>
add_link("soil", "mamInsect") # |>
#add_link("soil", "birdInsect")
plot(trophic_df, shift=FALSE)
spcmdl_simple <- spacemodel(stack_habitat, trophic_df)
kernels <- list(
soil = NA,
#plant = NA,
#invert = NA,
mamHerb = NA,
mamInsect = NA#,
#birdInsect = NA
)
data("sf_micromammals")
sf_micromammals$trophic = ifelse(sf_micromammals$genus %in% c("crocidura", "sorex"), "insectivore", "herbivore")
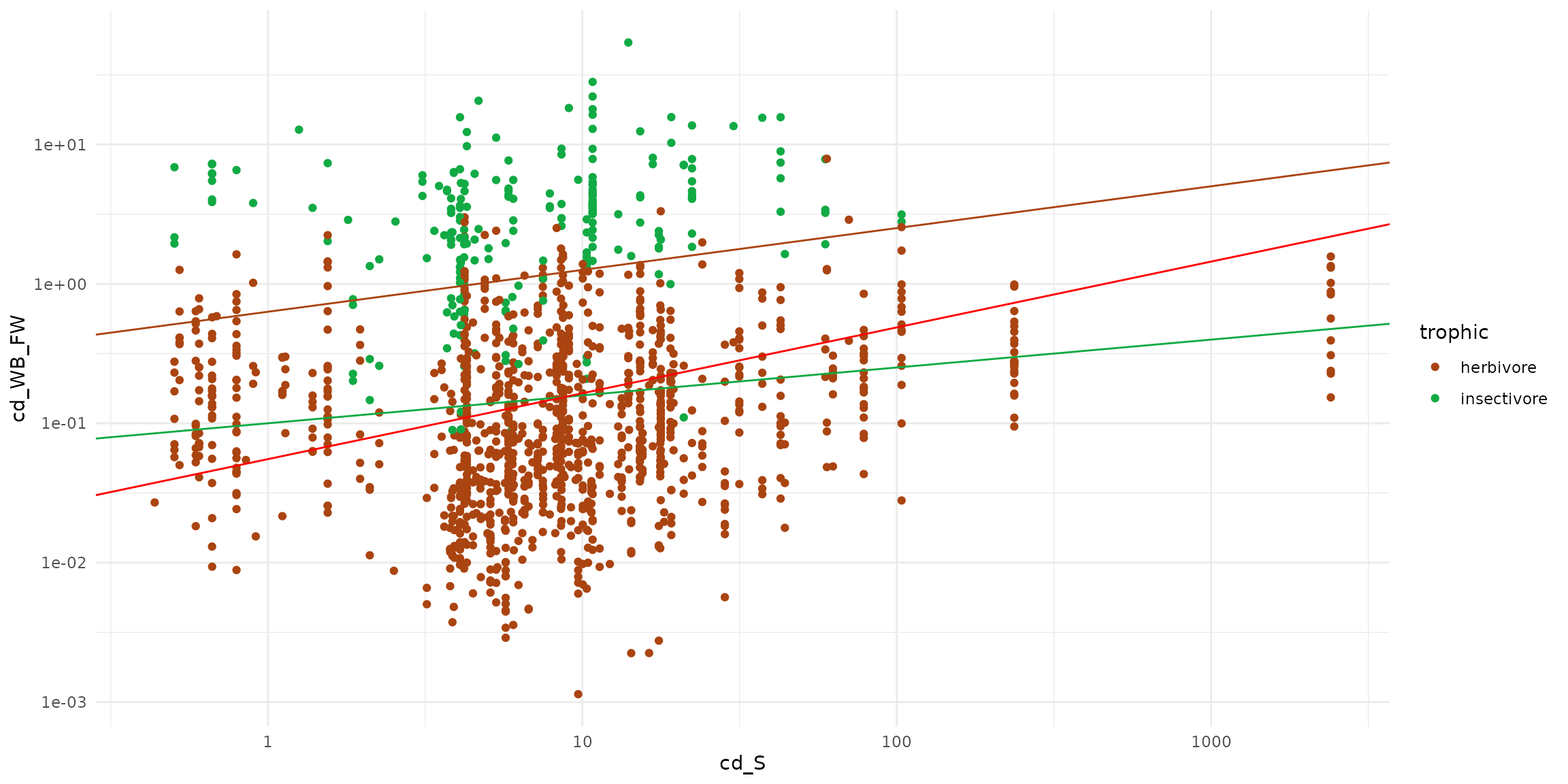
ggplot(data = sf_micromammals) +
theme_minimal() +
scale_x_log10() + scale_y_log10() +
scale_color_manual(values=c("#aa4411", "#11aa44")) +
geom_point(aes(x=cd_S, y=cd_WB_FW, color=trophic)) +
geom_abline(intercept= -0.2, slope=0.3, color = "#aa4411" ) +
geom_abline(intercept= -1, slope=0.2, color = "#11aa44" ) +
geom_abline(intercept= -1.2571, slope=0.4723, color = "red")
simple_intakes <- intake(spcmdl_simple,
"soil -> mamHerb" = ~ 10^(-1 + 0.2*x),
"soil -> mamInsect" = ~ 10^(-0.2 + 0.3*x),
default = 1 # for all other default is 1
)
spcmdl_transfer <- transfer(spcmdl_simple, kernels, simple_intakes)
terra::plot(spcmdl_transfer)
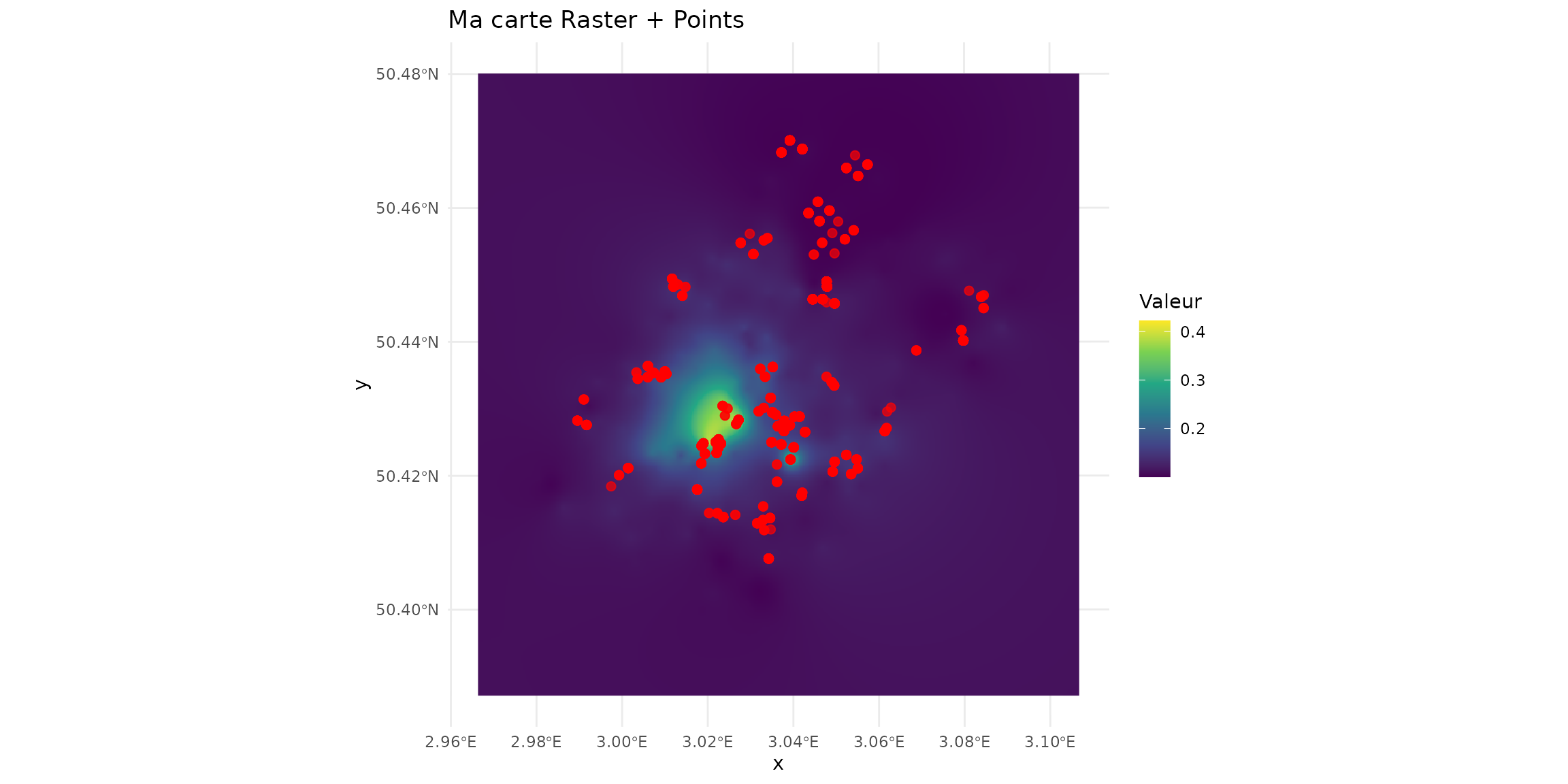
ptsInsect = sf_micromammals[sf_micromammals$trophic=="insectivore", ]
ptsHerb = sf_micromammals[sf_micromammals$trophic=="herbivore", ]
predict_insect <- terra::extract(spcmdl_transfer[["mamInsect"]], ptsInsect)
ptsInsect$predict <- predict_insect[, "mamInsect"]
predict_herb <- terra::extract(spcmdl_transfer[["mamHerb"]], ptsHerb)
ptsHerb$predict <- predict_herb[, "mamHerb"]
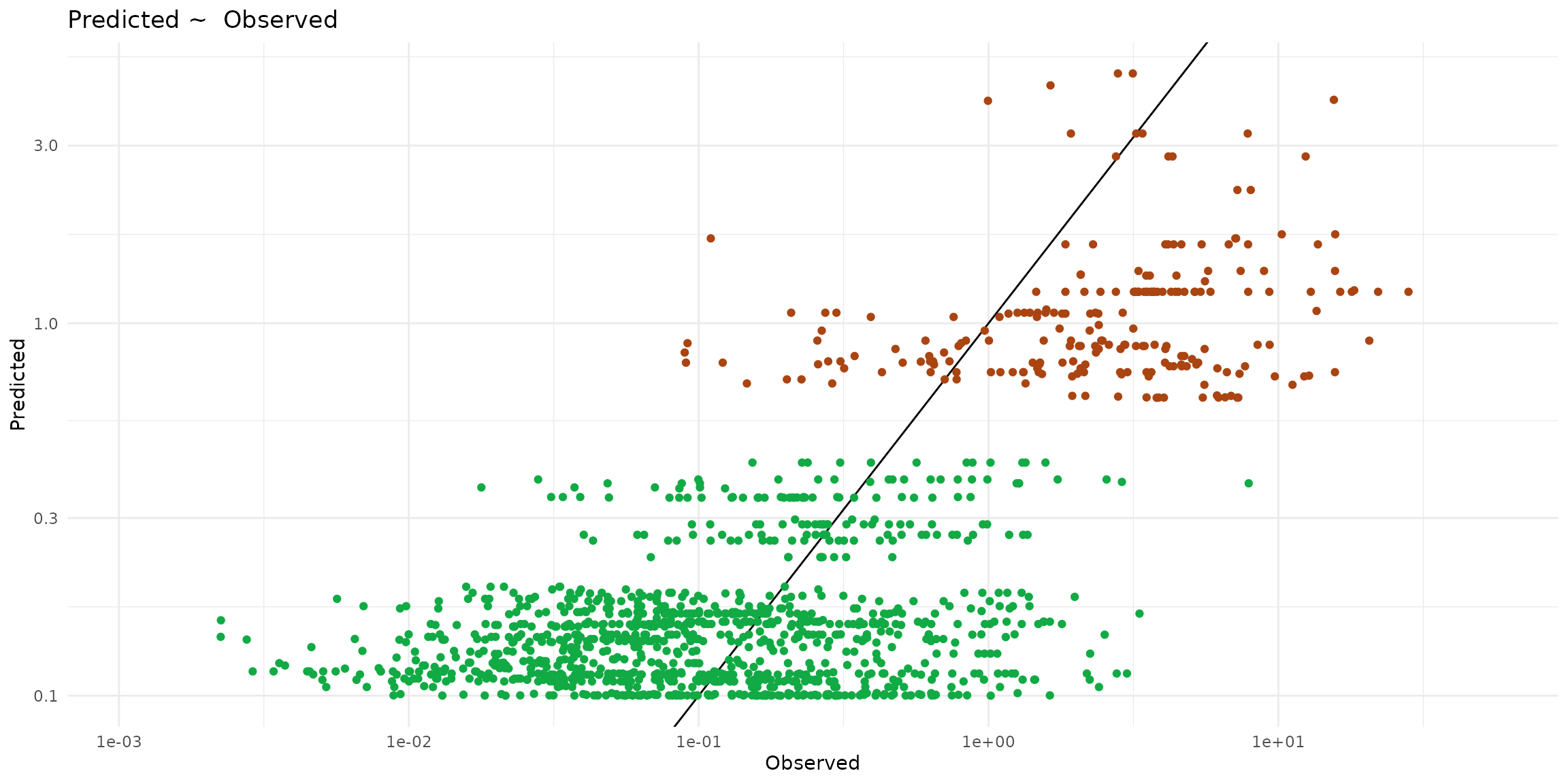
ggplot() +
theme_minimal() +
scale_x_log10() + scale_y_log10() +
labs(title="Predicted ~ Observed", x = "Observed", y = "Predicted") +
geom_abline(slope=1) +
geom_point(data = ptsInsect, color="#aa4411",
aes(x=cd_WB_FW, y=predict, color=trophic)) +
geom_point(data = ptsHerb, color="#11aa44",
aes(x=cd_WB_FW, y=predict, color=trophic))
#> Warning: Removed 7 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 111 rows containing missing values or values outside the scale range
#> (`geom_point()`).
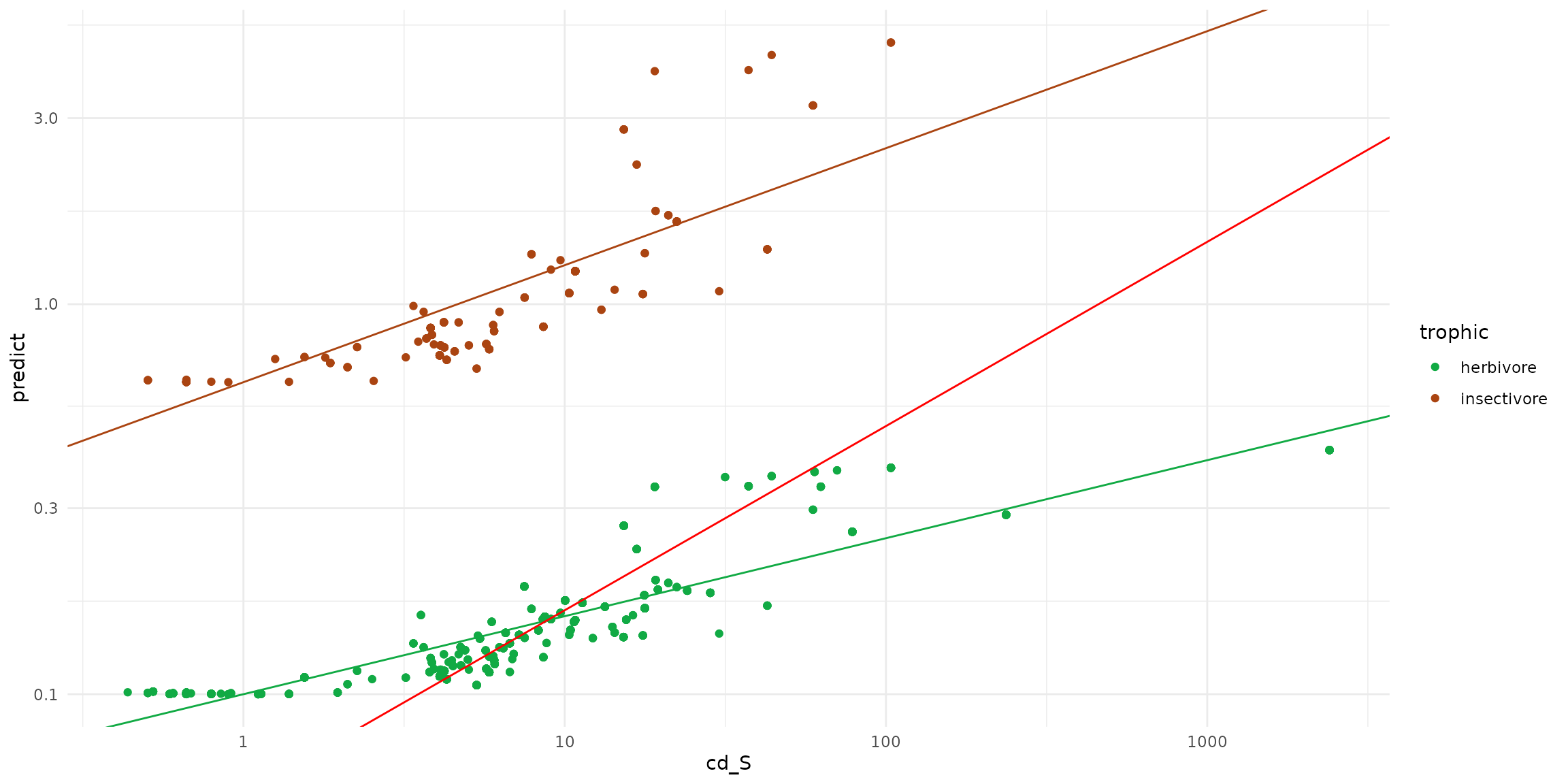
sf_predict = rbind(ptsInsect, ptsHerb)
ggplot(data = sf_predict) +
theme_minimal() +
scale_x_log10() + scale_y_log10() +
scale_color_manual(values=c("#11aa44", "#aa4411")) +
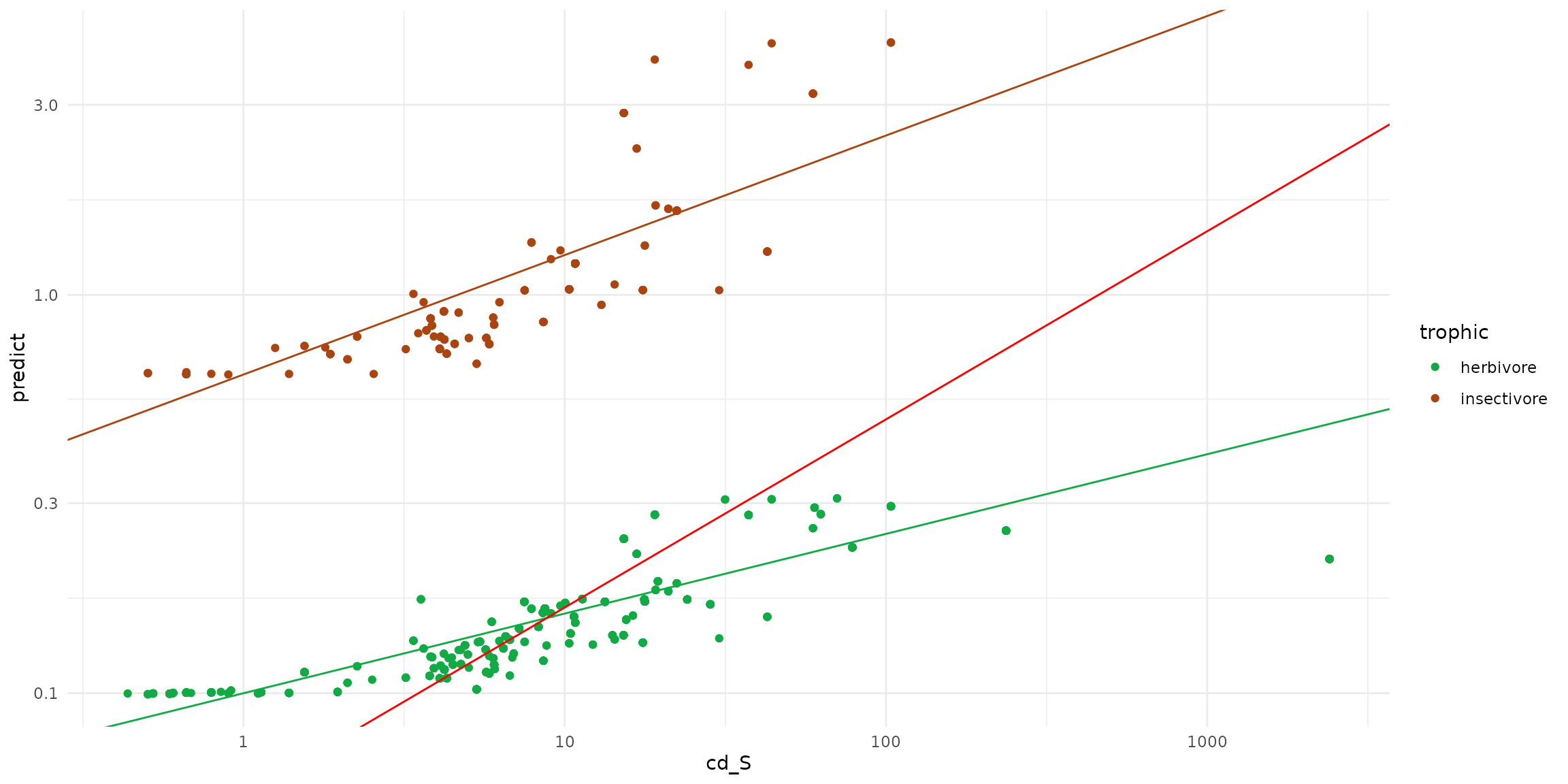
geom_point(aes(x=cd_S, y=predict, color=trophic)) +
geom_abline(intercept= -0.2, slope=0.3, color = "#aa4411" ) +
geom_abline(intercept= -1, slope=0.2, color = "#11aa44" ) +
geom_abline(intercept= -1.2571, slope=0.4723, color = "red")
#> Warning: Removed 118 rows containing missing values or values outside the scale range
#> (`geom_point()`).
A simple Soil - Target model with Dispersal
ground_cd <- load_raster_extdata("ground_concentration_cd_compressed.tif")
# names_hab = c("soil", "plant", "invert", "mamHerb", "mamInsect", "birdInsect")
names_hab = c("soil", "mamHerb", "mamInsect")
list_habitat <- lapply(names_hab, function(i) ground_cd)
stack_habitat <- raster_stack(list_habitat, names_hab)
terra::plot(stack_habitat)
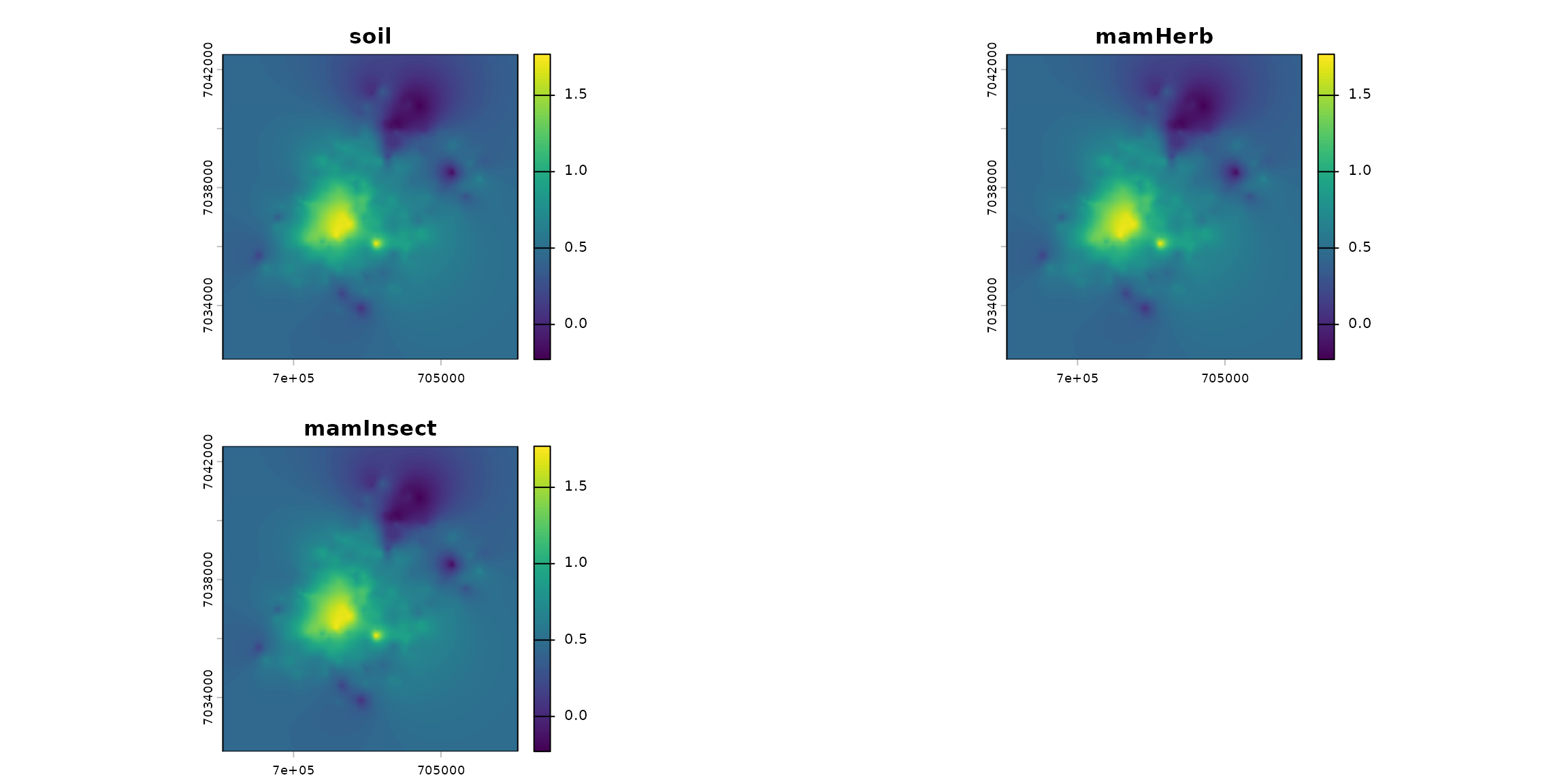
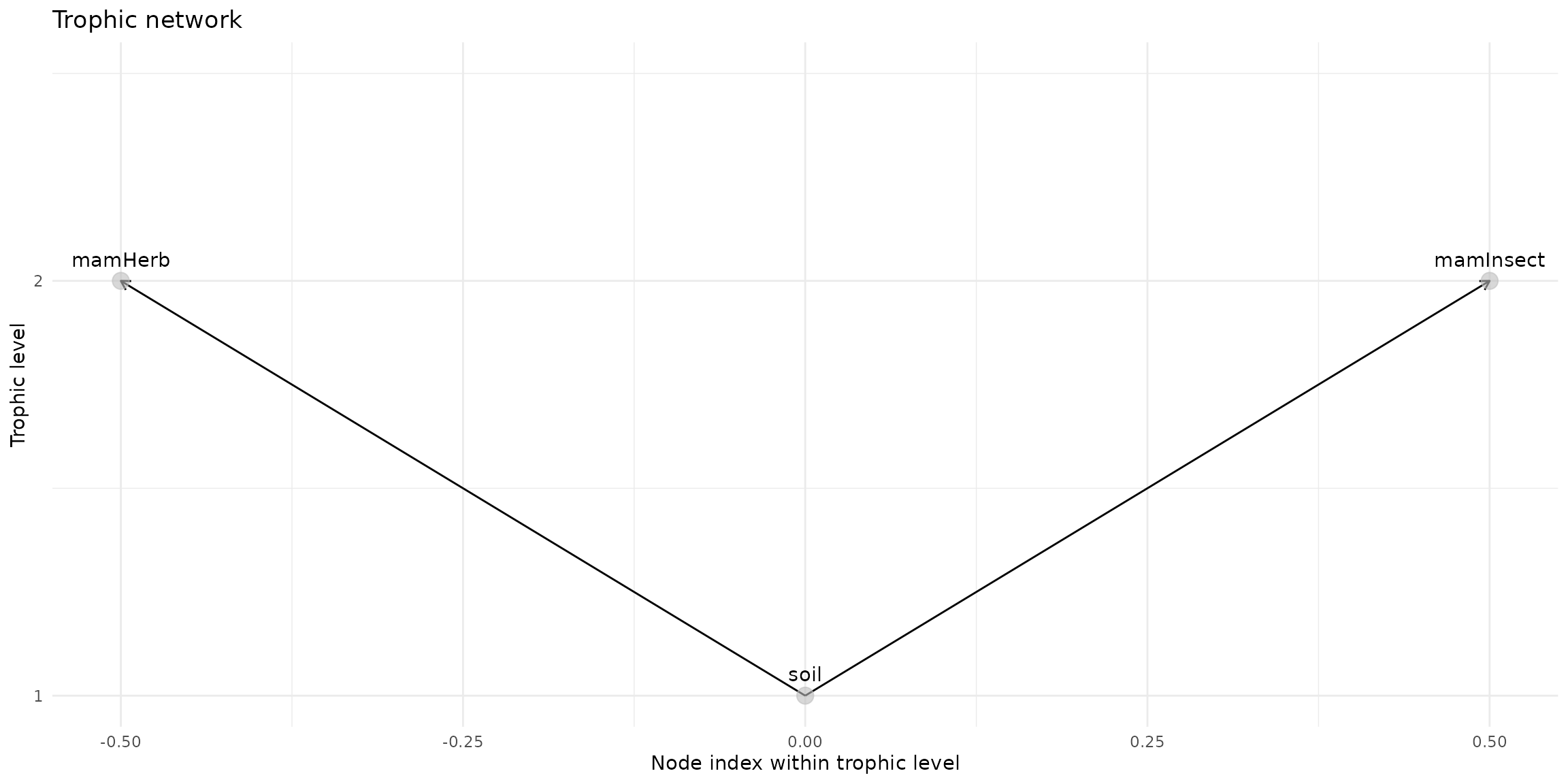
trophic_df <- trophic() |>
#add_link("soil", "plant") |>
#add_link("soil", "invert") |>
add_link("soil", "mamHerb") |>
add_link("soil", "mamInsect") # |>
#add_link("soil", "birdInsect")
plot(trophic_df, shift=FALSE)
spcmdl_simple <- spacemodel(stack_habitat, trophic_df)
k_mamInsect <- compute_kernel(radius=100, GSD=25, size_std=1.5)
k_mamHerb <- compute_kernel(radius=500, GSD=25, size_std=1.5)
kernels <- list(
soil = NA,
#plant = NA,
#invert = NA,
mamHerb = k_mamHerb,
mamInsect = k_mamInsect#,
#birdInsect = NA
)
simple_intakes <- intake(spcmdl_simple,
"soil -> mamHerb" = ~ 10^(-1 + 0.2*x),
"soil -> mamInsect" = ~ 10^(-0.2 + 0.3*x),
default = 1 # for all other default is 1
)
spcmdl_transfer <- transfer(spcmdl_simple, kernels, simple_intakes)
terra::plot(spcmdl_transfer)
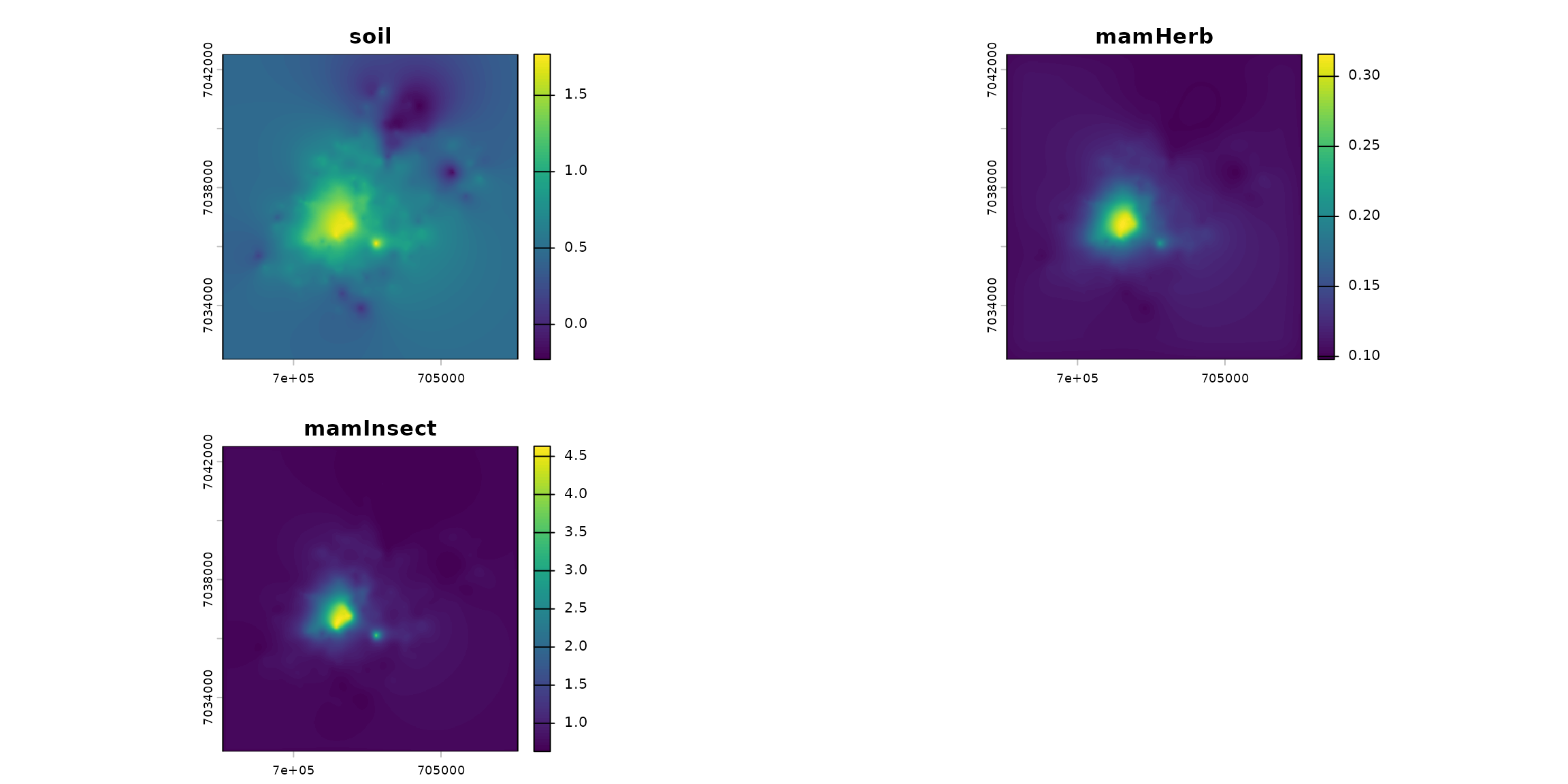
ptsInsect = sf_micromammals[sf_micromammals$trophic=="insectivore", ]
ptsHerb = sf_micromammals[sf_micromammals$trophic=="herbivore", ]
predict_insect <- terra::extract(spcmdl_transfer[["mamInsect"]], ptsInsect)
ptsInsect$predict <- predict_insect[, "mamInsect"]
predict_herb <- terra::extract(spcmdl_transfer[["mamHerb"]], ptsHerb)
ptsHerb$predict <- predict_herb[, "mamHerb"]
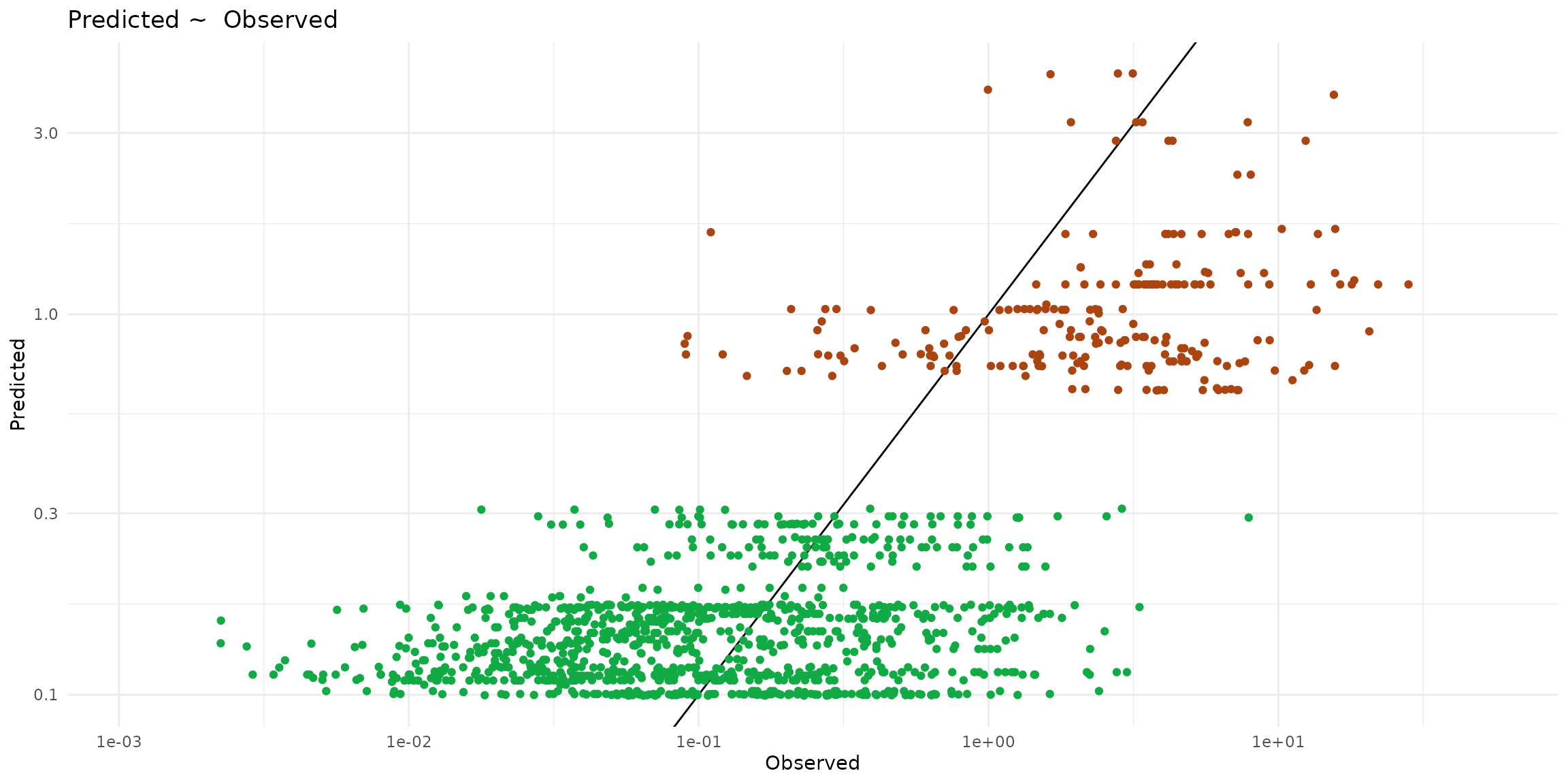
ggplot() +
theme_minimal() +
scale_x_log10() + scale_y_log10() +
labs(title="Predicted ~ Observed", x = "Observed", y = "Predicted") +
geom_abline(slope=1) +
geom_point(data = ptsInsect, color="#aa4411",
aes(x=cd_WB_FW, y=predict, color=trophic)) +
geom_point(data = ptsHerb, color="#11aa44",
aes(x=cd_WB_FW, y=predict, color=trophic))
#> Warning: Removed 7 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 111 rows containing missing values or values outside the scale range
#> (`geom_point()`).
sf_predict = rbind(ptsInsect, ptsHerb)
ggplot(data = sf_predict) +
theme_minimal() +
scale_x_log10() + scale_y_log10() +
scale_color_manual(values=c("#11aa44", "#aa4411")) +
geom_point(aes(x=cd_S, y=predict, color=trophic)) +
geom_abline(intercept= -0.2, slope=0.3, color = "#aa4411" ) +
geom_abline(intercept= -1, slope=0.2, color = "#11aa44" ) +
geom_abline(intercept= -1.2571, slope=0.4723, color = "red")
#> Warning: Removed 118 rows containing missing values or values outside the scale range
#> (`geom_point()`).